Visualise data
Adam Sparks and Ihsan Khaliq
2021-07-16
a01_Visualise_Data.RmdLoad libraries
library("readxl")
library("cowplot")
library("ggplot2")
library("ggpubr")
library("grDevices")
library("dplyr")
library("lubridate")
library("clifro")
library("viridis")
library("showtext")
library("readr")
library("here")
library("patchwork")
library("classInt")
library("extrafont")
extrafont::loadfonts()Import data
dat <-
read_excel(
system.file("extdata", "SpatioTemporalSpreadData_N.xlsx",
package = "spatiotemporaldynamics"),
sheet = 1
)Examine data
str(dat)## tibble [1,800 × 18] (S3: tbl_df/tbl/data.frame)
## $ location : chr [1:1800] "Billa Billa" "Billa Billa" "Billa Billa" "Billa Billa" ...
## $ assessment_date : POSIXct[1:1800], format: "2020-07-02" "2020-07-02" ...
## $ assessment_number: num [1:1800] 1 1 1 1 1 1 1 1 1 1 ...
## $ plot_number : num [1:1800] 1 1 1 1 1 1 1 1 1 1 ...
## $ distance : num [1:1800] 0 9 9 9 9 9 9 9 9 6 ...
## $ quadrat : chr [1:1800] "F" "N9" "NE9" "E9" ...
## $ direction : chr [1:1800] "All" "North" "NorthEast" "East" ...
## $ infected_plants : num [1:1800] 0 0 0 0 0 0 0 0 0 0 ...
## $ total_plants : num [1:1800] 36 48 27 57 53 41 39 31 36 54 ...
## $ incidence : num [1:1800] 0 0 0 0 0 0 0 0 0 0 ...
## $ min_temp : num [1:1800] 3.99 3.99 3.99 3.99 3.99 ...
## $ max_temp : num [1:1800] 20 20 20 20 20 ...
## $ avg_temp : num [1:1800] 12 12 12 12 12 ...
## $ avg_wind_speed : num [1:1800] 1.52 1.52 1.52 1.52 1.52 ...
## $ total_rain : num [1:1800] 1 1 1 1 1 1 1 1 1 1 ...
## $ min_rh : num [1:1800] 35.2 35.2 35.2 35.2 35.2 ...
## $ max_rh : num [1:1800] 82.7 82.7 82.7 82.7 82.7 ...
## $ avg_rh : num [1:1800] 58.9 58.9 58.9 58.9 58.9 ...Convert variables to their correct classes
cols_1 <-
c(
"location",
"distance",
"plot_number",
"quadrat",
"min_temp",
"max_temp",
"min_rh",
"max_rh",
"avg_wind_speed",
"avg_rh",
"avg_temp",
"assessment_number",
"total_rain"
)
dat[cols_1] <- lapply(dat[cols_1], factor)
cols_2 <- c("infected_plants", "total_plants")
dat[cols_2] <- lapply(dat[cols_2], as.integer)
dat$assessment_date <- as.Date(dat$assessment_date)Re-check class
sapply(dat, class)## location assessment_date assessment_number plot_number
## "factor" "Date" "factor" "factor"
## distance quadrat direction infected_plants
## "factor" "factor" "character" "integer"
## total_plants incidence min_temp max_temp
## "integer" "numeric" "factor" "factor"
## avg_temp avg_wind_speed total_rain min_rh
## "factor" "factor" "factor" "factor"
## max_rh avg_rh
## "factor" "factor"Kernel denisty plots
Kernel density plot to visualise data distribution as by distance & location
ggplot(dat, aes(infected_plants)) +
geom_density() +
facet_grid(distance ~ location, labeller = label_both) +
theme_pubclean(base_family = "Arial Unicode MS")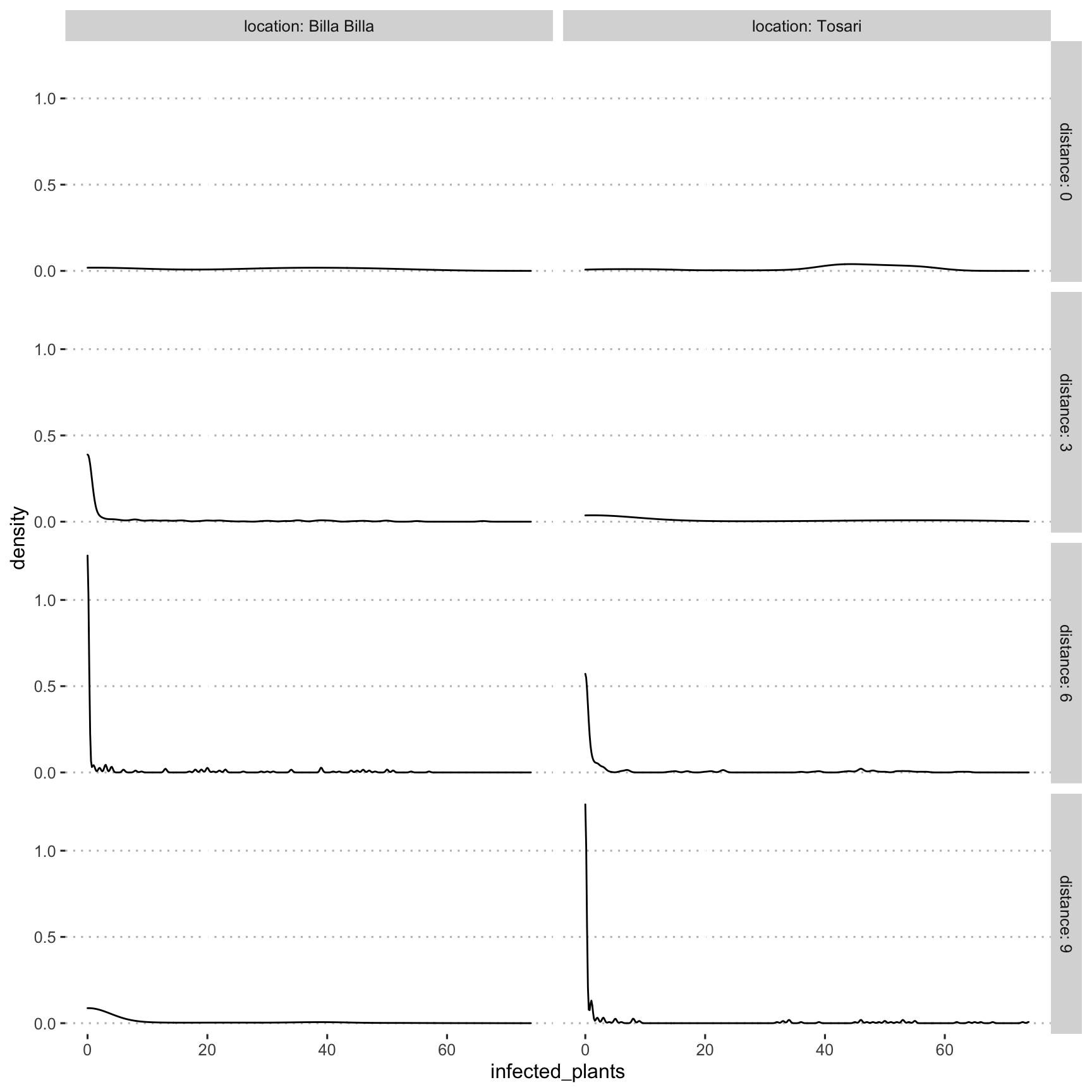
Kernel density plot to visualise overall data distribution
ggplot(dat, aes(x = infected_plants)) +
geom_density(fill= "steelblue", alpha = 0.7) +
geom_rug(alpha = 0.4) +
xlab("Infected plants count")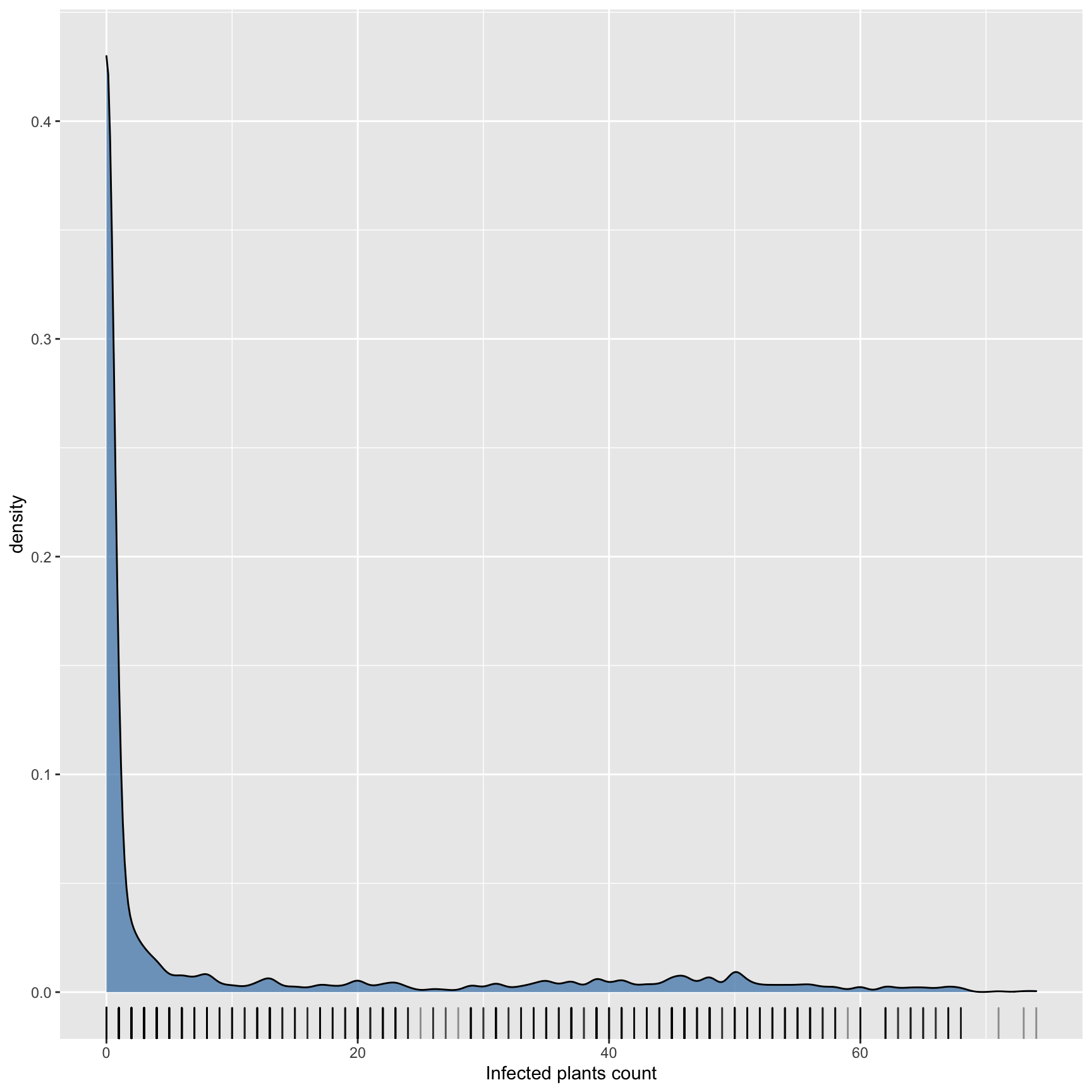
Line plot with median and max/min values
fig_1 <- ggplot(data = dat,
mapping = aes(x = assessment_number, y = infected_plants)) +
geom_pointrange(
stat = "summary",
fun.min = min,
fun.max = max,
fun = median
) +
stat_summary(fun = median,
geom = "line",
aes(group = location)) +
facet_grid(distance ~ location) +
xlab("Assessment number") +
ylab("Infected plants") +
theme_pubclean(base_family = "Arial Unicode MS")
fig_1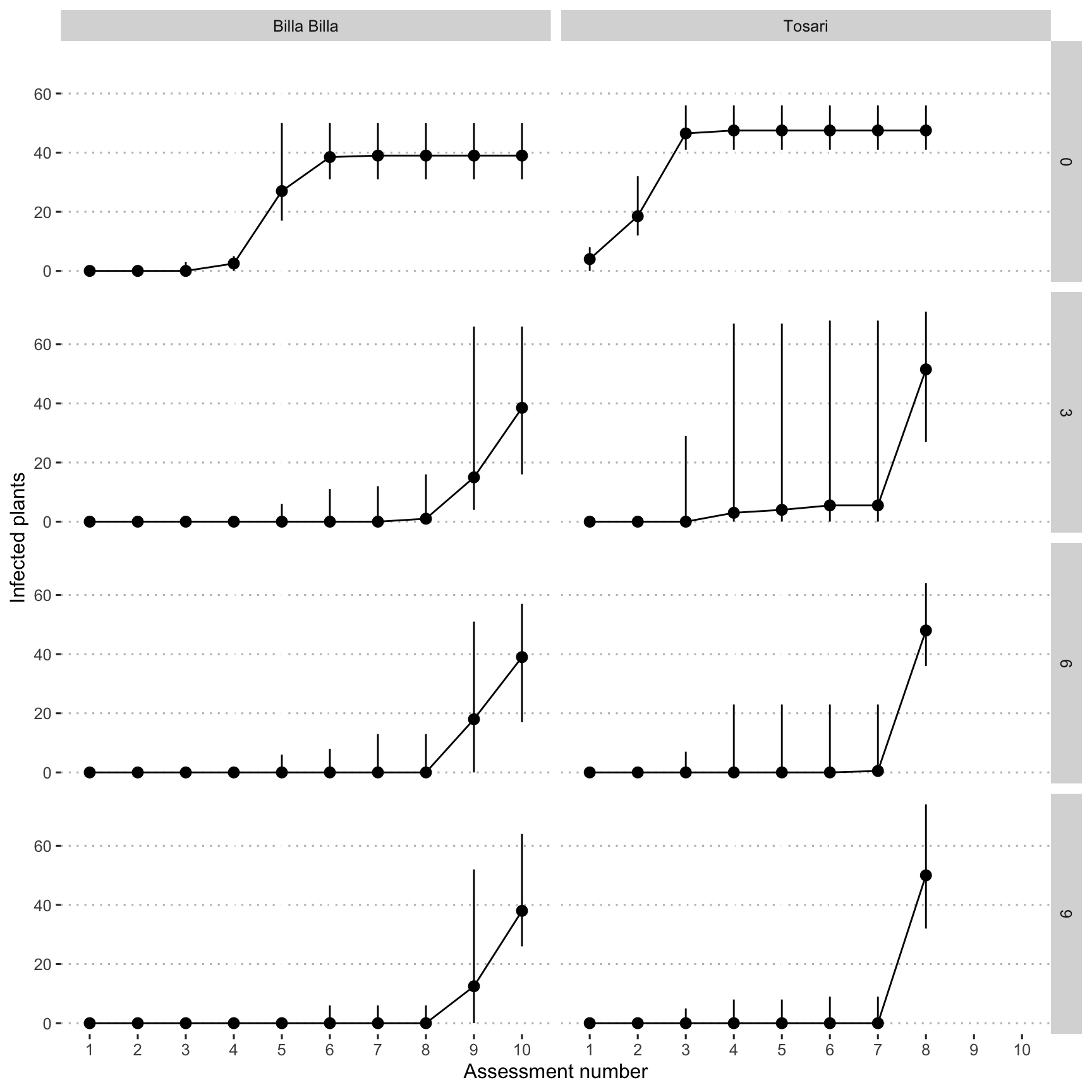
Wind rose
Import wind direction data and covert wind direction from text to degrees
# data munging
wind_direc_dat <-
read_csv(
system.file("extdata", "WindDirectionData.csv",
package = "spatiotemporaldynamics")
) %>%
mutate(
wind_direction_degrees = case_when(
wind_direction == "N" ~ "0",
wind_direction == "NbE" ~ "11.25",
wind_direction == "NNE" ~ "22.5",
wind_direction == "NEbN" ~ "33.75",
wind_direction == "NE" ~ "45",
wind_direction == "NEbE" ~ "56.25",
wind_direction == "ENE" ~ "67.5",
wind_direction == "EbN" ~ "73.5",
wind_direction == "E" ~ "90",
wind_direction == "EbS" ~ "101.2",
wind_direction == "ESE" ~ "112.5",
wind_direction == "SEbE" ~ "123.8",
wind_direction == "SE" ~ "135.1",
wind_direction == "SEbS" ~ "146.3",
wind_direction == "SSE" ~ "157.6",
wind_direction == "SbE" ~ "168.8",
wind_direction == "S" ~ "180",
wind_direction == "SbW" ~ "191.2",
wind_direction == "SSW" ~ "202.5",
wind_direction == "SWbS" ~ "213.8",
wind_direction == "SW" ~ "225",
wind_direction == "SWbW" ~ "236.2",
wind_direction == "WSW" ~ "247.5",
wind_direction == "WbS" ~ "258.8",
wind_direction == "W" ~ "270",
wind_direction == "WbN" ~ "281.2",
wind_direction == "WNW" ~ "292.5",
wind_direction == "NWbW" ~ "303.8",
wind_direction == "NW" ~ "315",
wind_direction == "NWbN" ~ "326.2",
wind_direction == "NNW" ~ "337.5",
wind_direction == "NbW" ~ "348.8",
TRUE ~ wind_direction
)
) %>%
mutate(wind_direction_degrees = as.numeric(wind_direction_degrees)) %>%
mutate(date = dmy(date))##
## ── Column specification ────────────────────────────────────────────────────────
## cols(
## date = col_character(),
## location = col_character(),
## wind_direction = col_character()
## )Join wind speed and wind direction data
# Import wind speed data
wind_speed_dat <-
read_csv(system.file("extdata", "WindSpeedData.csv",
package = "spatiotemporaldynamics")) %>%
mutate(date = as_date(dmy_hm(date)))##
## ── Column specification ────────────────────────────────────────────────────────
## cols(
## date = col_character(),
## location = col_character(),
## wind_speed = col_double()
## )
### Join wind speed and wind direction data
wind_dat <- left_join(wind_speed_dat, wind_direc_dat)## Joining, by = c("date", "location")Plot windrose over chickpea growing season
fig_2 <-
with(
wind_dat,
windrose(
wind_speed,
wind_direction_degrees,
location,
n_col = 2,
legend_title = "Wind speed (m/s)"
)
)
fig_2 <-
fig_2 +
scale_fill_viridis_d(name = "Wind Speed (m/s)", direction = -1) +
xlab("") +
theme_pubclean(base_family = "Arial Unicode MS")
fig_2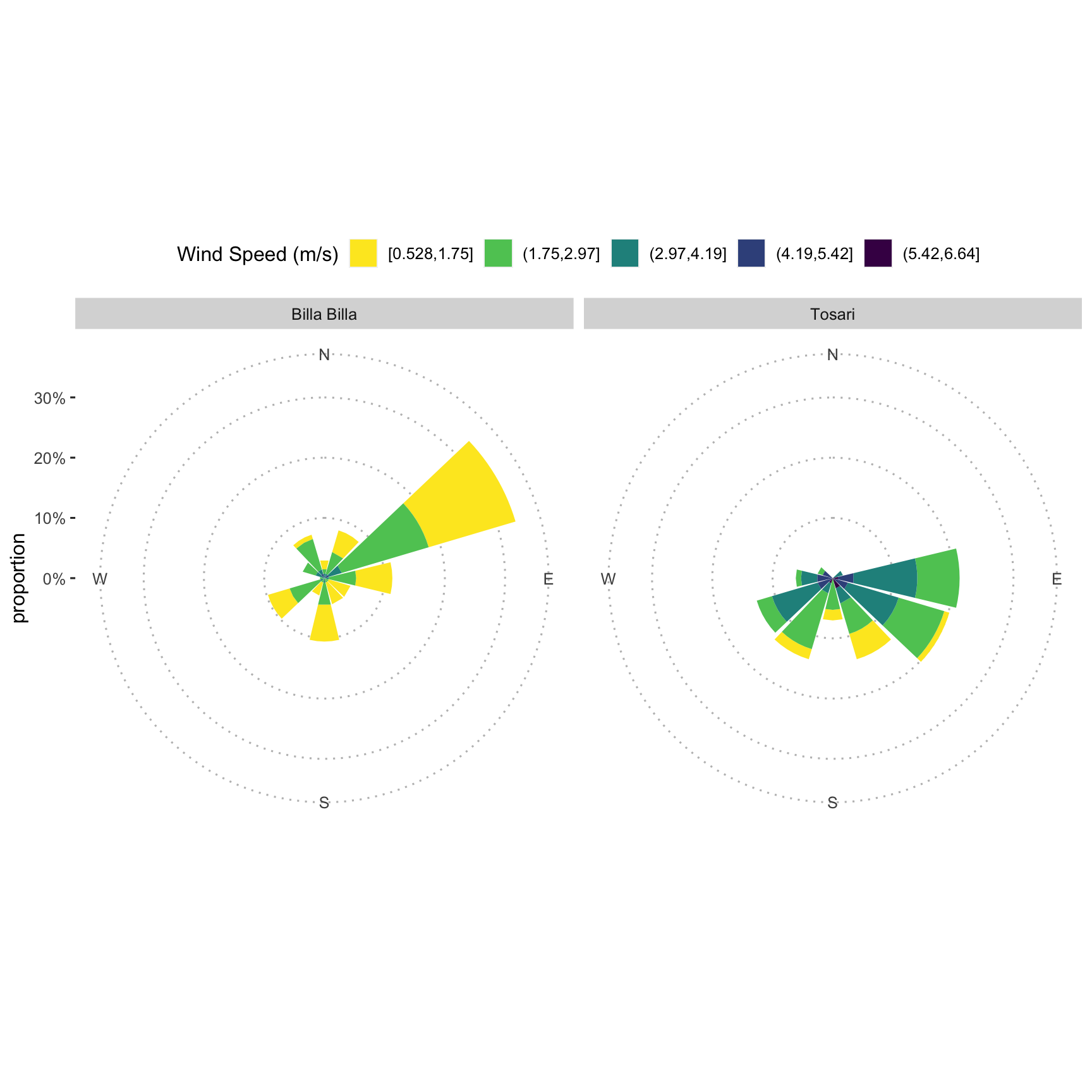
Data munging for ploting windrose by assessment number
# data munging
wind_dt <-
wind_dat %>%
mutate(
assessment_number =
case_when(
date <= "2020-07-02" & location == "Billa Billa" ~ 1,
date > "2020-07-02" &
date <= "2020-07-16" & location == "Billa Billa" ~ 2,
date > "2020-07-16" &
date <= "2020-07-30" & location == "Billa Billa" ~ 3,
date > "2020-07-30" &
date <= "2020-08-13" & location == "Billa Billa" ~ 4,
date > "2020-08-13" &
date <= "2020-08-26" & location == "Billa Billa" ~ 5,
date > "2020-08-26" &
date <= "2020-09-10" & location == "Billa Billa" ~ 6,
date > "2020-09-10" &
date <= "2020-09-24" & location == "Billa Billa" ~ 7,
date > "2020-09-24" &
date <= "2020-10-08" & location == "Billa Billa" ~ 8,
date > "2020-10-08" &
date <= "2020-10-22" & location == "Billa Billa" ~ 9,
date > "2020-10-22" &
date <= "2020-10-27" &
location == "Billa Billa" ~ 10,
date <= "2020-07-30" & location == "Tosari" ~ 1,
date > "2020-07-30" &
date <= "2020-08-14" & location == "Tosari" ~ 2,
date > "2020-08-14" &
date <= "2020-08-27" & location == "Tosari" ~ 3,
date > "2020-08-27" &
date <= "2020-09-10" & location == "Tosari" ~ 4,
date > "2020-09-10" &
date <= "2020-09-25" & location == "Tosari" ~ 5,
date > "2020-09-25" &
date <= "2020-10-09" & location == "Tosari" ~ 6,
date > "2020-10-09" &
date <= "2020-10-23" & location == "Tosari" ~ 7,
date > "2020-10-23" &
date <= "2020-11-05" & location == "Tosari" ~ 8
)
) %>%
mutate(assessment_number = as.factor(assessment_number))
Billa_Billa_wind_dt <- filter(wind_dt, location == "Billa Billa")
Tosari_wind_dt <- filter(wind_dt, location == "Tosari") %>%
droplevels()Plot windrose
# create breaks for the windroses
breaks <-
classIntervals(wind_dat$wind_speed, n = 4, style = "jenks")
# Billa Billa windrose
fig_3.1 <-
with(
Billa_Billa_wind_dt,
windrose(
wind_speed,
wind_direction_degrees,
facet = assessment_number,
n_col = 5,
legend_title = "Wind speed (m/s)",
speed_cuts = c(0, 1.4, 2.8, 4.2, 5.6, 7)
)
)
fig_3.1 <-
fig_3.1 +
scale_fill_viridis_d(name = "Wind Speed (m/s)",
direction = -1,
option = "cividis") +
scale_y_continuous(name = "Proportion (%)",
labels = c(0, 10, 20, 30, 40, 50, 60)) +
xlab("") +
theme_pubclean(base_family = "Arial Unicode MS") +
theme(
axis.ticks.length = unit(0, "mm"),
axis.line = element_blank(),
panel.spacing.x = unit(0, "lines"),
panel.spacing.y = unit(0, "lines"),
plot.margin = margin(0, 0, 0, 0, "cm")
)
# Tosari windrose
fig_3.2 <-
with(
Tosari_wind_dt,
windrose(
wind_speed,
wind_direction_degrees,
facet = assessment_number,
n_col = 5,
legend_title = "Wind speed (m/s)",
speed_cuts = c(0, 1.4, 2.8, 4.2, 5.6, 7)
)
)
fig_3.2 <-
fig_3.2 +
scale_fill_viridis_d(name = "Wind Speed (m/s)",
direction = -1,
option = "cividis") +
scale_y_continuous(name = "Proportion (%)",
labels = c(0, 10, 20, 30, 40, 50)) +
xlab("") +
theme_pubclean(base_family = "Arial Unicode MS") +
theme(
legend.position = "none",
axis.ticks.length = unit(0, "mm"),
axis.line = element_blank(),
panel.spacing.x = unit(0, "lines"),
panel.spacing.y = unit(0, "lines"),
plot.margin = margin(0, 0, 0, 0, "cm")
)
fig_3 <-
fig_3.1 / fig_3.2
fig_3 <-
fig_3 +
plot_annotation(tag_levels = "A") &
theme(plot.tag = element_text())
fig_3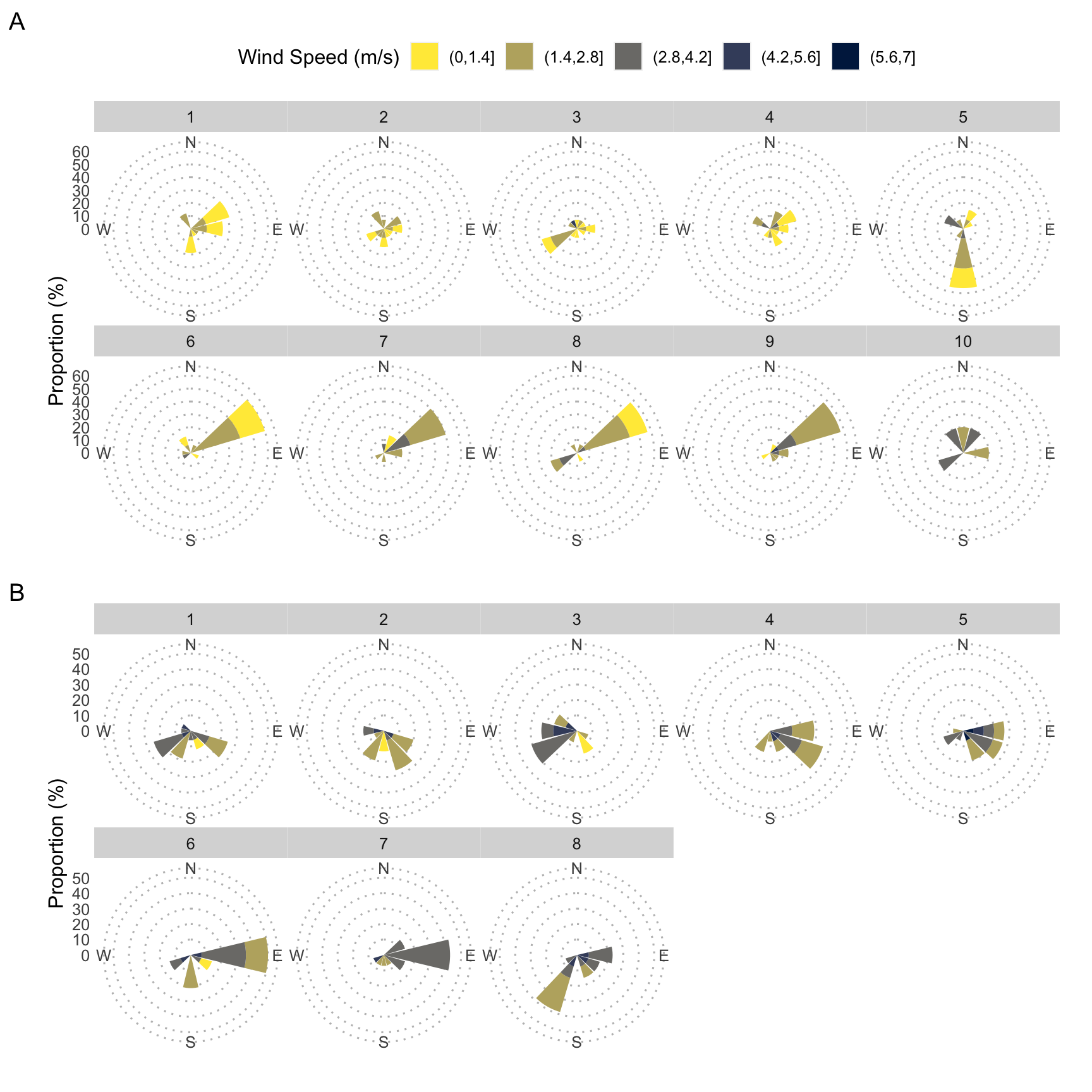
Colophon
## R version 4.1.0 (2021-05-18)
## Platform: x86_64-apple-darwin17.0 (64-bit)
## Running under: macOS Catalina 10.15.6
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRblas.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_AU.UTF-8/en_AU.UTF-8/en_AU.UTF-8/C/en_AU.UTF-8/en_AU.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] extrafont_0.17 classInt_0.4-3 patchwork_1.1.1 here_1.0.1
## [5] readr_1.4.0 showtext_0.9-2 showtextdb_3.0 sysfonts_0.8.3
## [9] viridis_0.6.1 viridisLite_0.4.0 clifro_3.2-5 lubridate_1.7.10
## [13] dplyr_1.0.7 ggpubr_0.4.0 ggplot2_3.3.5 cowplot_1.1.1
## [17] readxl_1.3.1
##
## loaded via a namespace (and not attached):
## [1] fs_1.5.0 RColorBrewer_1.1-2 httr_1.4.2 rprojroot_2.0.2
## [5] tools_4.1.0 backports_1.2.1 bslib_0.2.5.1 utf8_1.2.1
## [9] R6_2.5.0 KernSmooth_2.23-20 DBI_1.1.1 colorspace_2.0-2
## [13] withr_2.4.2 tidyselect_1.1.1 gridExtra_2.3 curl_4.3.2
## [17] compiler_4.1.0 extrafontdb_1.0 cli_3.0.0 rvest_1.0.0
## [21] xml2_1.3.2 desc_1.3.0 labeling_0.4.2 sass_0.4.0
## [25] scales_1.1.1 proxy_0.4-26 pkgdown_1.6.1 stringr_1.4.0
## [29] digest_0.6.27 foreign_0.8-81 rmarkdown_2.9 rio_0.5.27
## [33] pkgconfig_2.0.3 htmltools_0.5.1.1 highr_0.9 fastmap_1.1.0
## [37] rlang_0.4.11 rstudioapi_0.13 farver_2.1.0 jquerylib_0.1.4
## [41] generics_0.1.0 jsonlite_1.7.2 zip_2.2.0 car_3.0-11
## [45] magrittr_2.0.1 Rcpp_1.0.7 munsell_0.5.0 fansi_0.5.0
## [49] abind_1.4-5 lifecycle_1.0.0 stringi_1.6.2 yaml_2.2.1
## [53] carData_3.0-4 plyr_1.8.6 grid_4.1.0 forcats_0.5.1
## [57] crayon_1.4.1 haven_2.4.1 hms_1.1.0 knitr_1.33
## [61] pillar_1.6.1 ggsignif_0.6.2 reshape2_1.4.4 glue_1.4.2
## [65] evaluate_0.14 data.table_1.14.0 vctrs_0.3.8 Rttf2pt1_1.3.8
## [69] cellranger_1.1.0 gtable_0.3.0 purrr_0.3.4 tidyr_1.1.3
## [73] assertthat_0.2.1 cachem_1.0.5 xfun_0.24 openxlsx_4.2.4
## [77] broom_0.7.8 e1071_1.7-7 rstatix_0.7.0 class_7.3-19
## [81] tibble_3.1.2 memoise_2.0.0 ellipsis_0.3.2